
Introduction
Here we present the result you will obtain from the analysis of the sequences in the program's Demo Mode. They are useful to see what different matrices in EpiQuest-H see within the sequences, and which areas they define as hydrophilic or hydrophobic.

EpiQuest-B | EpiQuest-A | EpiQuest-IM | EpiQuest-C |EpiQuest-T | EpiQuest-H | EpiQuest-M | In Charge | EpiStat
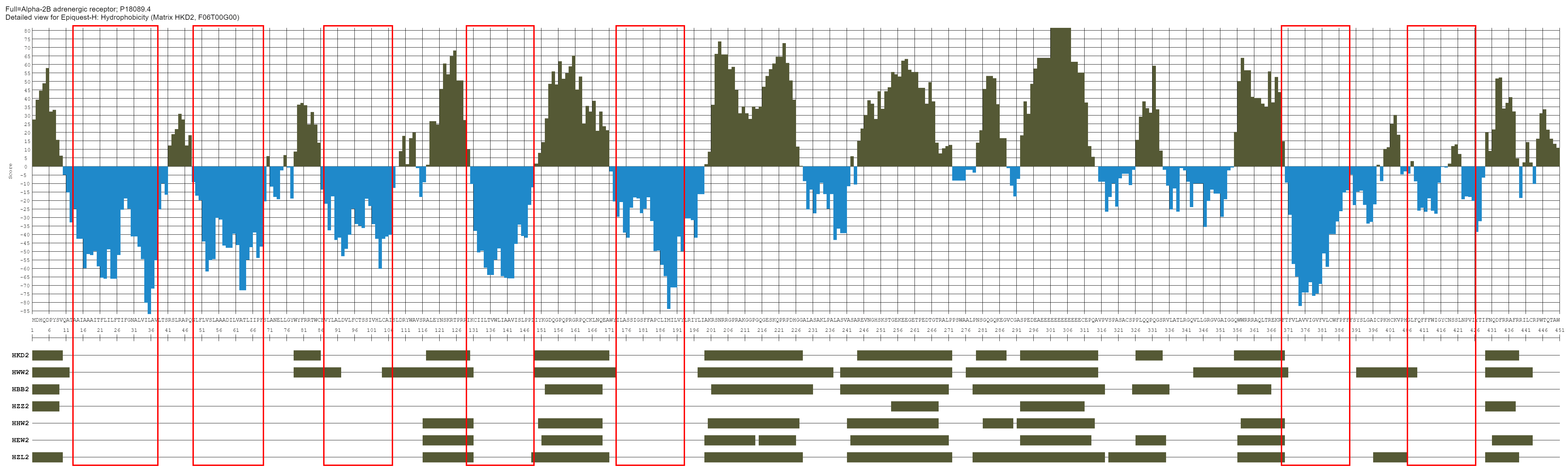
Alpha-2B adrenergic receptor
This molecule has 7 transmembrane domains (all indicated in red squares in the image below). You can see which algorithms define the TM domains correctly.
No less important is which of the algorithms defines the sequences of cytoplasmic and extracellular domains, and for what peaks there is a consensus between what are seen as hydrophilic sequences by different matrices.
Tetraspanin-18 [Homo sapiens]

Tetraspanin-18 has 4 transmembrane domains, indicated on the scheme as red boxes.
Please note which algorithms detect the sequences between these domains as hydrophilic, and for which sequences different matrices give a consensus.
Podoplanin [Homo sapiens]
This is the analysis of human podoplanin, a protein with one transmembrane domain (indicated by red box).
Please note that hydrophobic area surrounding the transmembrane domain is larger than the domain itself, and hydrophobicity profile cannot always be fully relied upon in establishing the transmembrane domain.
Additionally, please note how the various matrices differ in defining the hydrophilic domains, although there are clearly the sequences that are equally recognized by practically all matrices.

EpiQuest Suite and site www.epiquest.co.uk belongs to Aptum Biologics Ltd.
EpiQuest® is a registered Trademark of Aptum Biologics Ltd.
© 2018, 2020, 2021 Aptum Biologics Ltd.



